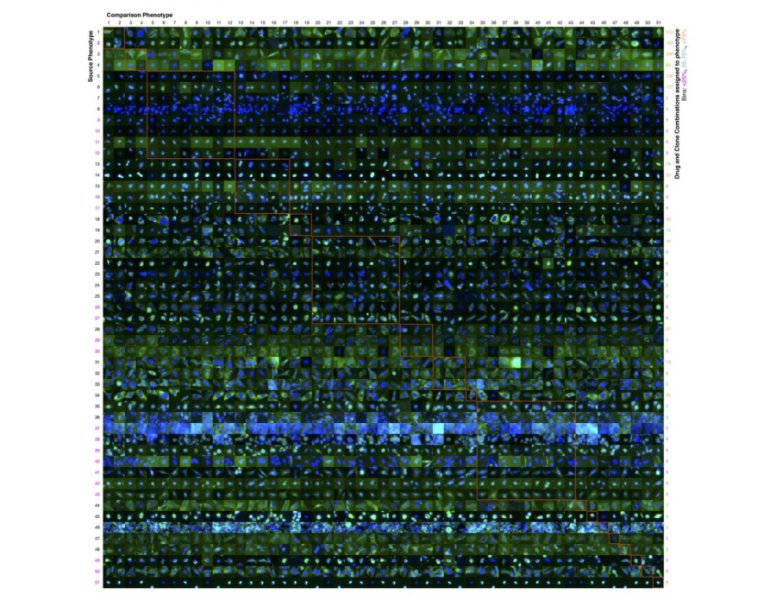
NEW YORK — Researchers, including an Indian American scientist, from Carnegie Mellon University (CMU) have created the first robotically driven experimentation system to determine the effects of a large number of drugs on many proteins, reducing the number of necessary experiments by 70%.
“Biomedical scientists have invested a lot of effort into making it easier to perform numerous experiments quickly and cheaply,” said lead author Armaghan Naik from Carnegie Mellon University’s computational biology department.
“However, we simply cannot perform an experiment for every possible combination of biological conditions, such as genetic mutation and cell type. Researchers have, therefore, had to choose a few conditions or targets to test exhaustively, or pick experiments themselves. The question is which experiments do you pick,” Naik added.
For this, Naik’s team previously described the application of a machine learning approach called “active learning.”
This involves a computer repeatedly choosing which experiments to do in order to learn efficiently from the patterns it observed in the data.
While their approach had only been tested using synthetic or previously acquired data, the team’s current model builds on this by letting the computer choose which experiments to do.
The experiments were then carried out using liquid-handling robots and an automated microscope. As the system progressively performed the experiments, it identified more phenotypes and more patterns in how sets of proteins were affected by sets of drugs.
The model was recently presented in the journal eLife.





Be the first to comment